BEST Services Help
Jason Vertrees
Date: 2005-01-07
The intent of this document is to provide and introduction to and assistance with the Hilser Lab's BEST (Biology using Ensemble-based Structural Thermodynamics) online services, hereafter ``BEST''.
Contents
0.1 Quick Start
To begin using the system the following is recommend:
- Register: (http://best.utmb.edu/BEST/registration.php).
- Sign in.
- Create a new workspace in your home directory.
- Upload a PDB from your computer, into your newly made workspace.
- Submit (any) Jobs.
- Entropy Weighting
- Complete Ensemble
- Stability Constants
- Check out your results.
0.2 Detailed Introduction
0.2.1 Registration
New user registration is completed via BEST's online registration page (http://best.utmb.edu/BEST/registration.php). Job submission status reports are emailed to the user, therefore valid email addresses are needed for registration. Authorization is immediate, once registered you may enter the BEST site (http://best.utmb.edu/).
The secure hypertext transport protocol (https) is not utilized at this time; please don't use any passwords that you may use in your lab or on other machines as transferring information through the standard http protocol allows for 3rd party interception of your data. (This goes for all sites, not just BEST.)
0.2.2 Using the Web Site
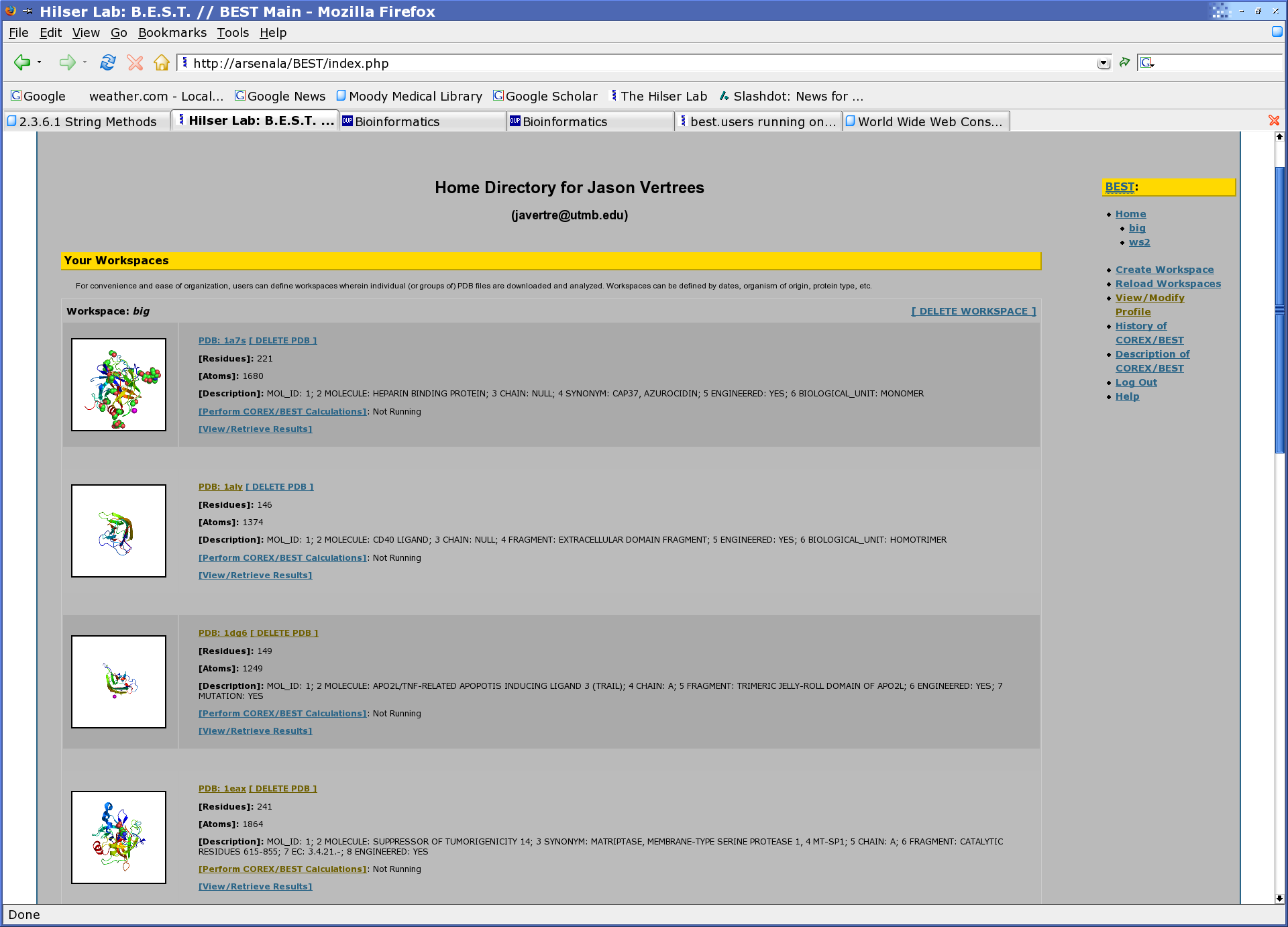
BEST's online presence is best experienced with any W3C (World Wide Web Consortium) standards compliant browser and is therefore platform independent. The suggested browser is any recent version of the Mozilla Foundation's Mozilla or FireFox web browser. See (http://www.mozilla.org) for more information. The web interface (v0.1b) is organized as a home directory with workspaces and within each workspace multiple PDB files (with results). An image of an example user's home directory is shown below.
0.2.2.1 Logging In
Once successfully registered, please visit (http://best.utmb.edu/BEST) to login.
0.2.2.2 Logging Out
To logout, click the ``Log Out'' link on the navigation bar at the right side of any BEST page.
0.2.3 Workspaces
Workspaces are convenient storage spaces with multiple proteins: store all similar proteins in one, or all proteins for a given experiment in another. One must have a workspace in which to upload PDBs. Workspace names must be alphanumeric (underscores '_' are also allowed in workspace names).
0.2.3.1 Adding a Workspace
One's first task is to create a workspace. The ``Create Workspace'' link will assist you in creating a new workspace. You may have multiple workspace in your user area.
0.2.3.2 Workspace Refreshing & Pending Results
Once a calculation is completed, the user's online profile is updated and the results may be accessed through the ``View/Review Results'' link for the corresponding PDB. Some results may not be caught by the web browser so a ``Reload Workspace'' link is provided letting the user manually & forcefully reload his or her workspace.
0.2.4 Upload a PDB
All BEST calculations are based off a PDB file. To upload a PDB to the BEST server, select the ``Upload New PDB to Workspace'' link at the bottom of the appropriate workspace. The upload process may take a while depending on i) your internet connection & size of PDB and ii) the load on the server. Resubmitting or selecting the upload button more than once will only lengthen the delay: please be patient. Once uploaded an image and protein specific information is shown in the user's home directory.
NOTE: PDB files with multiple models are not allowed. You must extract the header information and ONE model and place those into one file for uploading.
0.2.5 Submit Jobs
To enqueue ensemble-based calculations into our array of computers, please select a workspace and then a protein. Once chosen, click, ``Perform COREX/BEST Calculations''. This link leads you to a page with all appropriate forms for BEST ensemble calculations (with more calculations coming soon). Select the desired calculation, enter your data (default entries already there) and submit the values for processing. Jobs taking a long time - such as an ensemble - provide an estimated calculation time; shorter calculations - like entropy weighting - link directly to the results. Since, some calculations, like ``wsconf,'' ``dgover,'' and ``dgtemp'' are quick and some (ensemble generation) are possibly VERY long, some data may be missing from your profile if the browser reloads before the server writes the results to file.
0.2.5.1 Ensemble Generator
The ensemble generator is the base of most of BEST's calculations. Once a PDB is uploaded, it is first suggested that the user run an ensemble generator job.
0.2.5.2 Entropy Weighting
The next calculation suggested is the Entropy Weighting Factor.
0.2.5.3 Stability Constant Calculation
Once the above two calculations are complete, one my submit a stability constant calculation request. If the entropy weighting has not been calculated, a default value will be provided, otherwise the calculated entropy weighting will be provided. Note, the user may change the value to something other than the default or calculated entropy weighting.
0.2.6 Results
Each PDB in each workspace has its own set of results accessed via the ``View/Retrieve Results'' link next to the PDB profile. Some results are purely numerical, some possibly gigabytes of compressed data and others images, regardless, all results go here.
0.2.6.1 Personal Profile
The interface that handles personal information (name, email address, password) is easily updated by logging in and selecting the ``Modify Profile'' link.
0.2.6.2 Password Retrieval
Since we don't know your passwords (they are encrypted in our database) we can't send it to you. You may however request your password be reset to a random password which is then emailed to you. You may then login to change your password to something more manageable.